Q&A 21 How do you cluster data using hierarchical clustering or DBSCAN?
21.1 Explanation
While K-means assumes clusters are spherical and requires a preset k, other clustering methods offer different advantages:
- Hierarchical Clustering: Builds nested clusters based on distance — great for dendrograms and discovering cluster structure
- DBSCAN: Groups points based on density — ideal for detecting clusters of arbitrary shape and identifying outliers
We’ll demonstrate both using the gene_expression_with_clusters.csv dataset.
21.2 Python Code
# Hierarchical Clustering and DBSCAN in Python
import pandas as pd
import matplotlib.pyplot as plt
from scipy.cluster.hierarchy import dendrogram, linkage
from sklearn.cluster import DBSCAN
from sklearn.preprocessing import StandardScaler
# Load dataset
df = pd.read_csv("data/gene_expression_with_clusters.csv")
# Drop non-numeric column
X = df.drop(columns=['SampleID'])
# Standardize features
X_scaled = StandardScaler().fit_transform(X)
# Hierarchical clustering
linked = linkage(X_scaled, method='ward')
plt.figure(figsize=(10, 6))
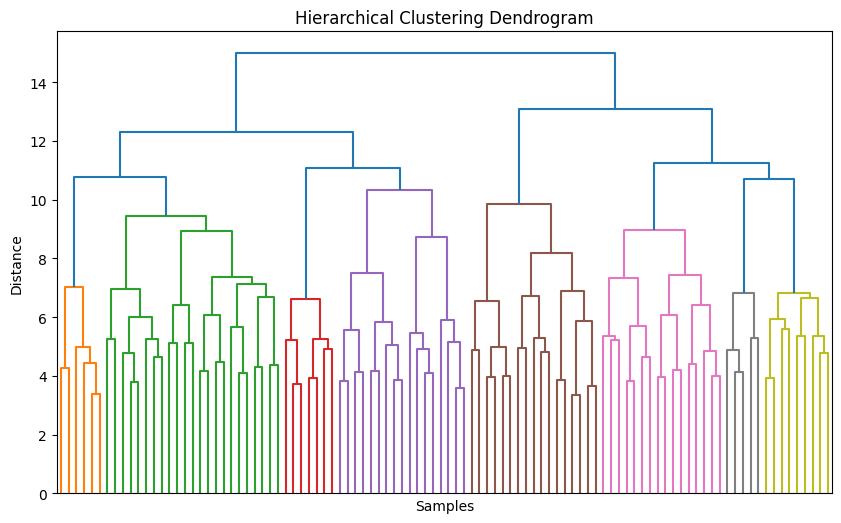
dendrogram(linked, no_labels=True)
plt.title("Hierarchical Clustering Dendrogram")
plt.xlabel("Samples")
plt.ylabel("Distance")
plt.show()
# DBSCAN
db = DBSCAN(eps=2, min_samples=5).fit(X_scaled)
df['DBSCAN_Cluster'] = db.labels_
# Visualize DBSCAN clusters (optional: with PCA)
from sklearn.decomposition import PCA
X_pca = PCA(n_components=2).fit_transform(X_scaled)
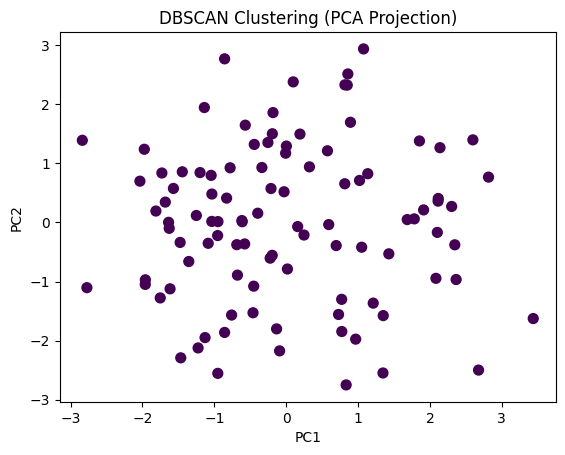
plt.scatter(X_pca[:, 0], X_pca[:, 1], c=df['DBSCAN_Cluster'], cmap='viridis', s=50)
plt.title("DBSCAN Clustering (PCA Projection)")
plt.xlabel("PC1")
plt.ylabel("PC2")
plt.show()
21.3 R Code
# Hierarchical Clustering and DBSCAN in R
library(readr)
library(dplyr)
library(ggplot2)
library(cluster)
library(dbscan)
library(factoextra)
# Load dataset
df <- read_csv("data/gene_expression_with_clusters.csv")
# Keep numeric columns
X <- df %>% select(-SampleID)
# Scale data
X_scaled <- scale(X)
# Hierarchical clustering
hc <- hclust(dist(X_scaled), method = "ward.D2")
plot(hc, main = "Hierarchical Clustering Dendrogram", xlab = "", sub = "")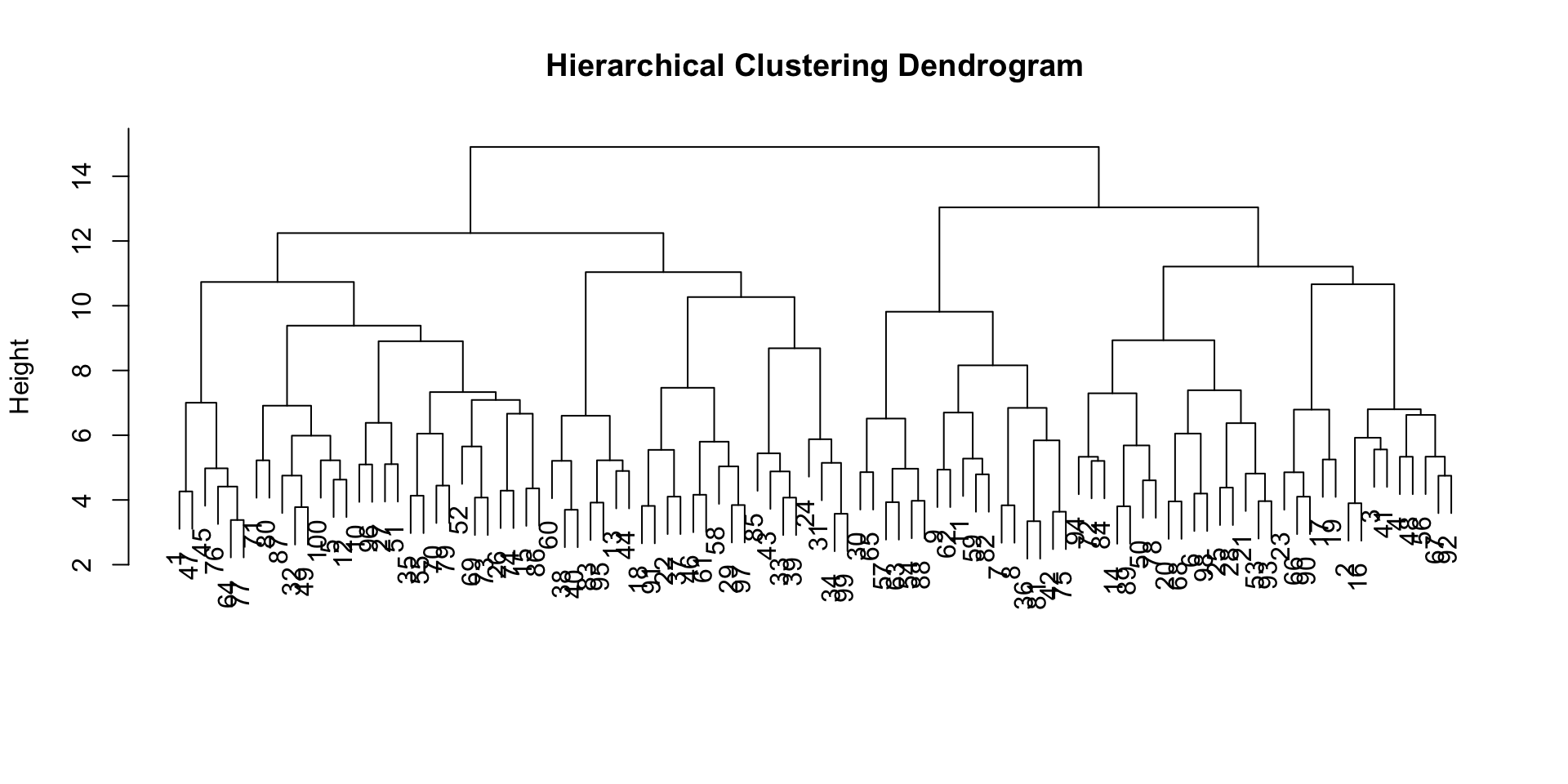
# DBSCAN
set.seed(42)
db <- dbscan(X_scaled, eps = 2, minPts = 5)
# PCA for visualization
pca <- prcomp(X_scaled)
pca_df <- as.data.frame(pca$x[, 1:2])
pca_df$cluster <- factor(db$cluster)
ggplot(pca_df, aes(x = PC1, y = PC2, color = cluster)) +
geom_point(size = 2) +
labs(title = "DBSCAN Clustering (PCA Projection)\n") +
theme_minimal()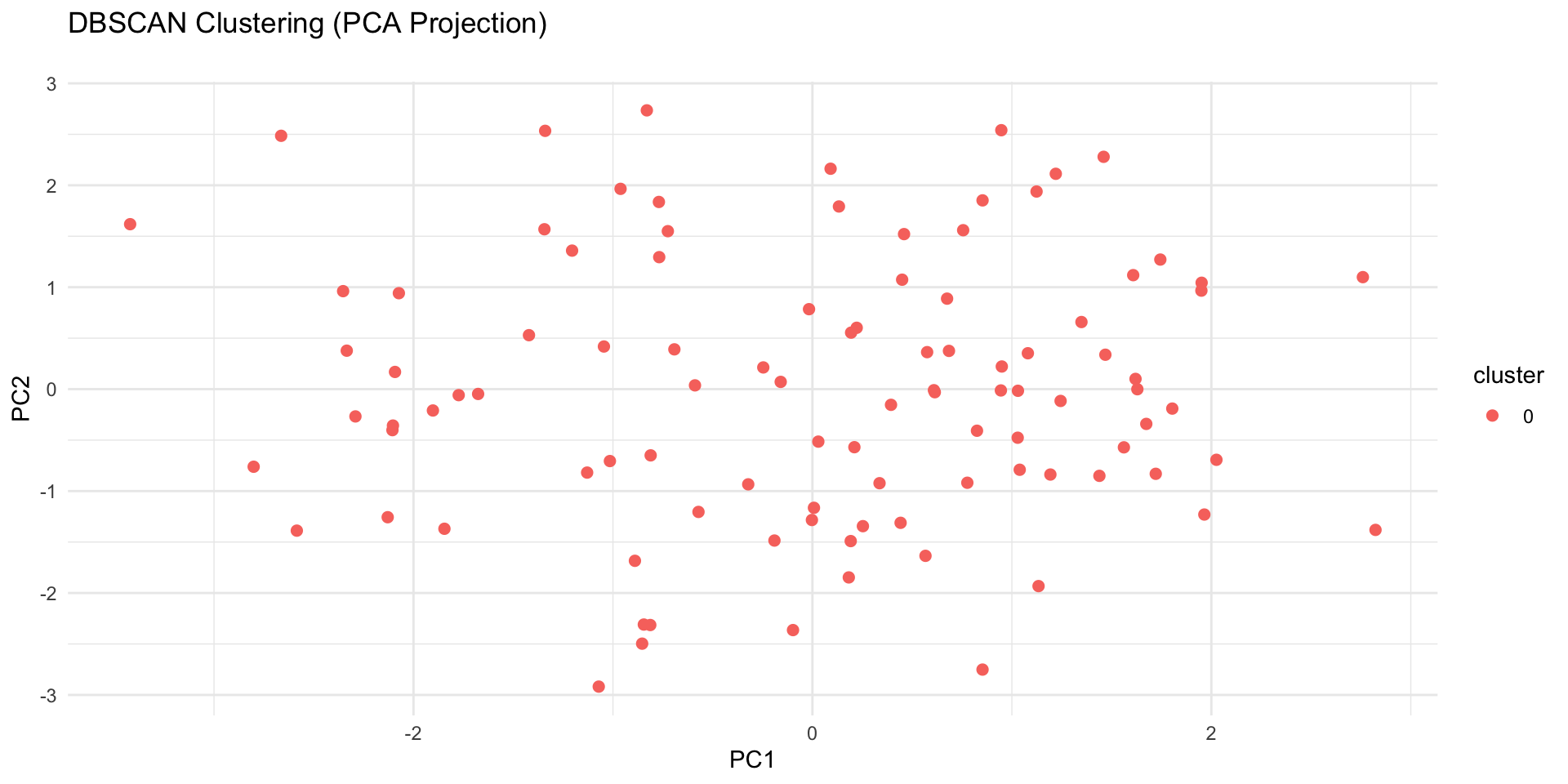
✅ Takeaway: Hierarchical clustering helps uncover structure without predefining clusters, while DBSCAN excels at finding complex shapes and outliers. When K-means isn’t enough, these methods reveal deeper patterns in your data.