Q&A 26 How do you create a heatmap to compare model performance across metrics?
26.1 Explanation
Once you’ve trained multiple models, comparing their performance across multiple metrics — like accuracy, precision, recall, F1-score, and AUC — provides a deeper understanding of their strengths and weaknesses.
Instead of viewing one metric at a time, a heatmap helps summarize all key metrics in one chart, allowing for quicker and more informed decisions when choosing a model.
26.2 Python Code
# Compare models using performance metrics heatmap (Python) — safe version
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score, roc_auc_score
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.svm import SVC
from sklearn.naive_bayes import GaussianNB
# Load and prepare Titanic data
df = pd.read_csv("data/titanic.csv")
df = df.dropna(subset=["Age", "Fare", "Embarked", "Sex", "Survived"])
# Make a copy to avoid SettingWithCopyWarning
X = df[["Pclass", "Age", "Fare"]].copy()
# Encode categorical variables safely using .loc
X.loc[:, "Sex"] = LabelEncoder().fit_transform(df["Sex"])
X.loc[:, "Embarked"] = LabelEncoder().fit_transform(df["Embarked"])
y = df["Survived"]
# Train-test split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=42)
# Models to compare
models = {
"LogReg": LogisticRegression(solver="liblinear"),
"RandomForest": RandomForestClassifier(n_estimators=100, random_state=42),
"SVM": SVC(probability=True, gamma="auto", random_state=42),
"NaiveBayes": GaussianNB()
}
# Evaluate and collect metrics
metrics_list = []
for name, model in models.items():
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
y_prob = model.predict_proba(X_test)[:, 1]
metrics_list.append({
"Model": name,
"Accuracy": accuracy_score(y_test, y_pred),
"Precision": precision_score(y_test, y_pred),
"Recall": recall_score(y_test, y_pred),
"F1": f1_score(y_test, y_pred),
"AUC": roc_auc_score(y_test, y_prob)
})
# Create DataFrame from metrics
metrics_df = pd.DataFrame(metrics_list).set_index("Model")
# Plot heatmap
plt.figure(figsize=(8, 5))
sns.heatmap(metrics_df, annot=True, fmt=".2f", cmap="YlGnBu")
plt.title("Model Performance Comparison")
plt.tight_layout()
plt.show()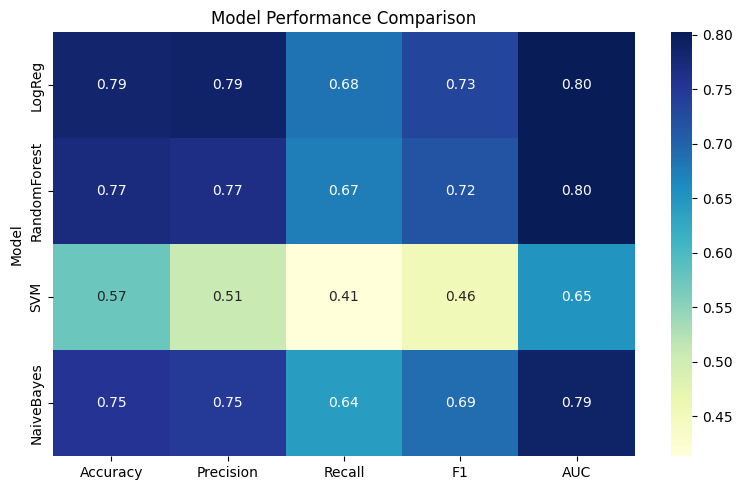
26.3 R Code
# Compare models using performance metrics heatmap (R)
library(readr)
library(dplyr)
library(tidyr)
library(caret)
library(pROC)
library(ggplot2)
library(reshape2)
# Load Titanic dataset
df <- read_csv("data/titanic.csv") %>%
drop_na(Age, Fare, Embarked, Sex, Survived)
# Prepare data
df <- df %>%
mutate(
Sex = as.factor(Sex),
Embarked = as.factor(Embarked),
Survived = as.factor(Survived)
)
# Features and labels
X <- df %>% dplyr::select(Pclass, Age, Fare, Sex, Embarked)
y <- df$Survived
# Train-test split
set.seed(42)
train_index <- createDataPartition(y, p = 0.7, list = FALSE)
train_data <- df[train_index, ]
test_data <- df[-train_index, ]
# Define formula with only clean columns
ml_formula <- Survived ~ Pclass + Age + Fare + Sex + Embarked
# Train models safely
models <- list(
LogReg = train(ml_formula, data = train_data, method = "glm", family = "binomial"),
RandomForest = train(ml_formula, data = train_data, method = "rf"),
SVM = train(ml_formula, data = train_data, method = "svmRadial"),
NaiveBayes = train(ml_formula, data = train_data, method = "naive_bayes")
)
# Metric evaluation function
get_metrics <- function(model, test_data) {
pred <- predict(model, test_data)
prob_df <- tryCatch({
predict(model, test_data, type = "prob")
}, error = function(e) {
warning(paste("Skipping prob for model due to:", e$message))
return(NULL)
})
obs <- test_data$Survived
if (!is.null(prob_df) && "Yes" %in% colnames(prob_df)) {
prob <- prob_df[["Yes"]]
auc_val <- tryCatch({
roc(obs, prob)$auc
}, error = function(e) {
warning(paste("AUC failed for model due to:", e$message))
return(NA)
})
} else {
prob <- rep(NA, length(obs))
auc_val <- NA
}
data.frame(
Accuracy = mean(pred == obs),
Precision = tryCatch(posPredValue(pred, obs, positive = "Yes"), error = function(e) NA),
Recall = tryCatch(sensitivity(pred, obs, positive = "Yes"), error = function(e) NA),
F1 = tryCatch(F_meas(pred, obs, positive = "Yes"), error = function(e) NA),
AUC = auc_val
)
}
# Collect all model metrics
metric_list <- lapply(models, get_metrics, test_data = test_data)
metric_df <- bind_rows(metric_list, .id = "Model")
# Plot heatmap
metric_long <- melt(metric_df, id.vars = "Model")
ggplot(metric_long, aes(x = variable, y = Model, fill = value)) +
geom_tile(color = "white") +
geom_text(aes(label = sprintf("%.2f", value)), size = 3) +
scale_fill_gradient(low = "yellow", high = "red") +
labs(title = "Model Performance Comparison", x = "Metric", y = "Model") +
theme_minimal()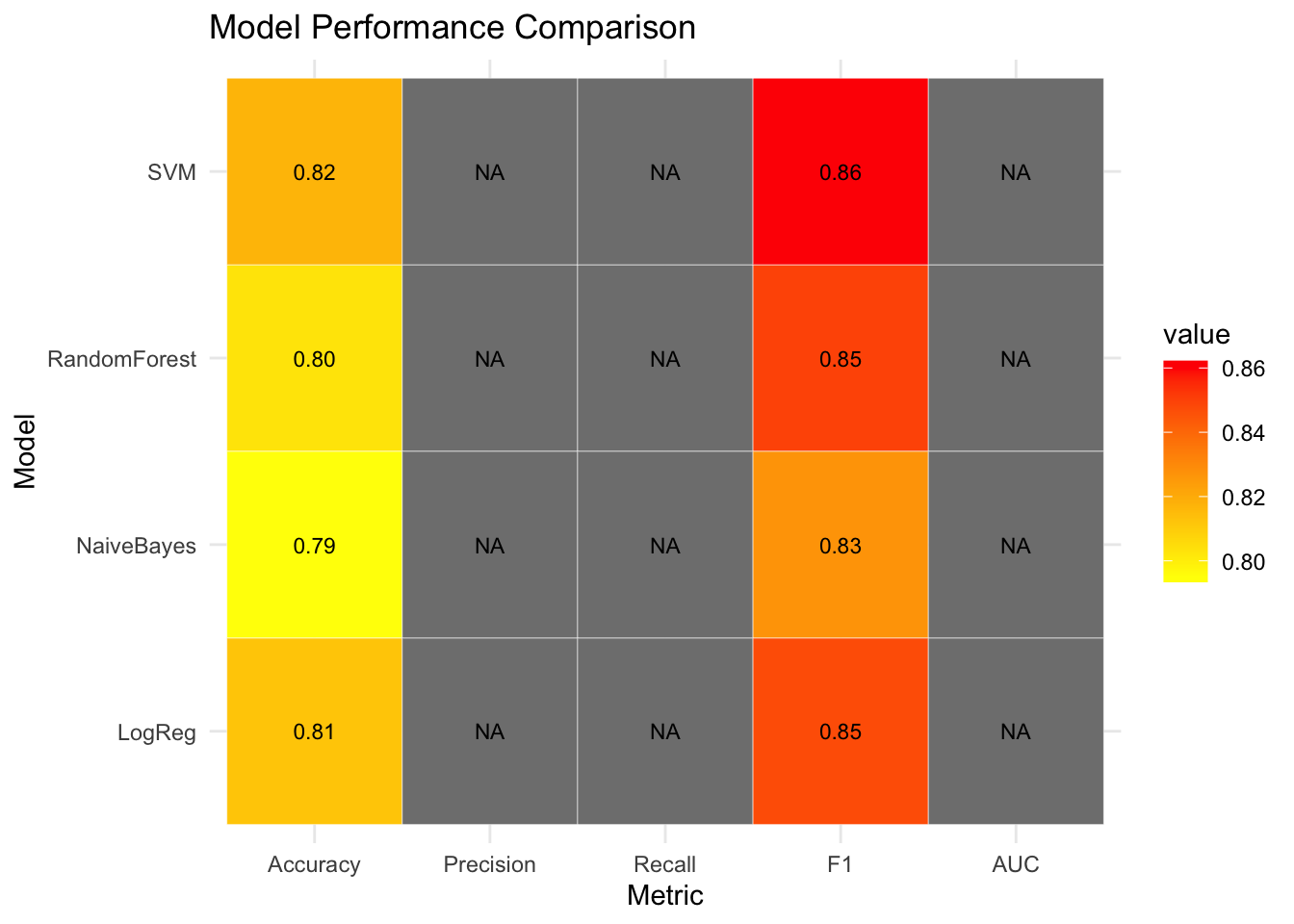
✅ Takeaway: Comparing multiple models in one place lets you make evidence-based decisions — not just based on accuracy, but also on precision, recall, F1, and AUC. A heatmap helps visually spot strengths and weaknesses across classifiers.